WHO Training
Dr. Beverly Egyir, WHO's Dr. Appiah-Korang Labi and Dr. Bright Adu led a five-day workshop at the Noguchi Memorial Institute for Medical Research that taught participants how to use whole-genome sequencing (WGS) for antimicrobial resistance (AMR) surveillance. It improved diagnostic and public health capabilities by combining theory, practical lab work, and data analysis.
REPORT ON WHO TRAINING WORKSHOP
WHOLE-GENOME SEQUENCING AND SURVEILLANCE OF ANTIMICROBIAL RESISTANCE IN BACTERIA
HOST INSTITUTION
NOGUCHI MEMORIAL INSTITUTE FOR MEDICAL RESEARCH (NMIMR), UNIVERSITY OF GHANA, (UG)
<
DAY ONE (6TH JUNE, 2022)
Introductions and Welcome Address.
Whole-genome sequencing (WGS) methods and applications were taught to participants from different Ghanaian labs at the Noguchi Memorial Institute. WHO's Dr. Appiah-Korang Labi and Dr. Beverly Egyir stressed the importance of using WGS to increase capacity for antimicrobial resistance (AMR) surveillance. The curriculum comprised facilities tours, hands-on lab work, and theoretical instruction.
Theory Session.
AMR surveillance, its worldwide effects, and the value of genotypic rather than phenotypic approaches for detecting resistance genes, monitoring outbreaks, and assessing virulence were all covered by Dr. Beverly Egyir. In addition to highlighting acquired and intrinsic resistance, she described WGS as a vital tool for AMR research and connected it to initiatives like Fleming/SeqAfrica and the SDGs.
Dr. Bright Adu discussed next-generation sequencing (NGS), including its history, technologies (Oxford Nanopore MinION, Illumina MiSeq), and molecular biology applications. He focused on factors like cost, time, and accuracy as well as short vs. long-read sequencing.
While Mr. Christian Owusu-Nyantakyi demonstrated DNA quantification techniques such as Nanodrop, qPCR (gold standard), Qubit, and Bioanalyzer, Ms. Felicia Owusu led DNA extraction methods using Qiagen kits. After that, participants took part in group activities and practical assignments.



 Practical Session
Practical Session
Using the Qiagen Kit, participants extracted bacterial DNA, which was then measured using the Qubit fluorometer and Nanodrop2000 spectrophotometer. Working with one gram-positive and one gram-negative bacterium, each team discovered that lysozyme is necessary for gram-positive DNA extraction because of their thicker peptidoglycan coatings. Participants were walked through the DNA extraction and quantification procedures step-by-step by facilitators. A Q&A and debates on the subjects discussed marked the end of the day.

 DAY 2 (7TH JUNE, 2022)
Theory Session
DAY 2 (7TH JUNE, 2022)
Theory Session
Hawa Ahmed delivered the day's overview, educating attendees on the lessons learned from earlier lectures, films, and hands-on activities. There have been concerns expressed regarding the preference for phenotypic AMR identification over the genotypic approach, which is the gold standard for resistance tracking and detection.
During his presentation of the Oxford Nanopore Technology workflow, Mr. Grebstad Rabbi Amuasi covered flow cell priming, loading libraries, library preparation, and nanopore theory. With an emphasis on the latter, which requires a 400ng concentration and may operate for 48–72 hours, he described the ligation and quick barcoding techniques. Prior to the practical session, participants saw videos.

 Practical Session
Practical Session
The participants were split into two groups: one was taught to Linux by Mr. Christian Owusu-Nyantakyi and Mr. Bright Agbodzi, while the other group used the quick barcoding kit to produce libraries for sequencing on the MinION platform under the supervision of Mr. Grebstad Rabbi Amuasi and Quaneeta Muhktar.
The MinION group finished the sequencing method, measured DNA using the Qubit Fluorometer, normalized libraries, and learnt how to eliminate bubbles when loading flow cells. The Linux group studied programs like Trimmomatic for trimming, Fastqc for quality checks, and Unicycler for sequence assembly while practicing fundamental commands (such as making folders and navigating) and analyzing Fastq files.
In order to enhance the participants' learning, Dr. George Kwasi Hedidoh from WHO's Tricycle Project came to visit, offered encouragement, and participated in the hands-on activity.

 DAY 3 (8TH JUNE, 2022)
Theory Session
DAY 3 (8TH JUNE, 2022)
Theory Session
Many of the attendees expressed excitement about the new knowledge and its applicability as Dr. Beverly Egyir welcomed them and talked about their experiences. The group leader of Team P2 led a review of what had happened the day before.
Mr. Bright Agbodzi gave a presentation on the Illumina Miseq procedure, emphasizing qPCR, normalization, tagmentation, and fragmentation. He gave a thorough explanation of tagmentation, illustrating the process of adding adapters to DNA segments with videos and graphs.
Mr. Grebstad Rabbi Amuasi discussed qPCR and the Bioanalyzer for library quality checks. He clarified why KAPA library quantification is favored due to its excellent accuracy and underlined the significance of making sure libraries fall within the 400–600 bp size range. To reinforce learning, participants viewed films on the qPCR and Bioanalyzer procedures.

 Practical Session
Practical Session
Participants discussed theory and reagents with facilitators during hands-on sessions using the Bioanalyzer and qPCR, prior to beginning,
In his talk on normalization, pooling, and denaturation, Mr. Christian Owusu-Nyantakyi emphasized the necessity of single-stranded DNA for sequencing as well as the four processes of library normalization.
Speaking about the Illumina Miseq's flow cell, cartridge, and built-in software, Ms. Quaneeta Mokhtar described how to load a run on the device.
In order to prepare for the final-day presentations, Dr. Beverly Egyir led the participants in a discussion of their team assignments that were provided via email. The day was then concluded when teams divided up to evaluate their assignments.
 DAY 4 (9TH JUNE, 2022)
Theory Session
DAY 4 (9TH JUNE, 2022)
Theory Session
The summary of the previous day's events was mediated by a member of team N1. Dr. Beverly Egyir gave an overview of the day's schedule and the significance of each task. Two groups of participants were formed; one group went to the sequencing lab, while the other group got ready for the group presentations the next day.
Participants learnt about the Illumina Miseq platform, including its components, flow cell setup, cartridge kinds and capacities, reagent handling, and machine washing with molecular-grade water, in the sequencing lab, which was supervised by Mr. Amuasi and Ms. Mohktar. Additionally, they learned how to obtain data from the MinION platform, including determining coverage and readiness, and practiced moving data from the Illumina Miseq to a hard drive.

 Practical Session
Practical Session
The properties of FASTA files were the main emphasis of the participants' introduction to internet resources and data processing tools.
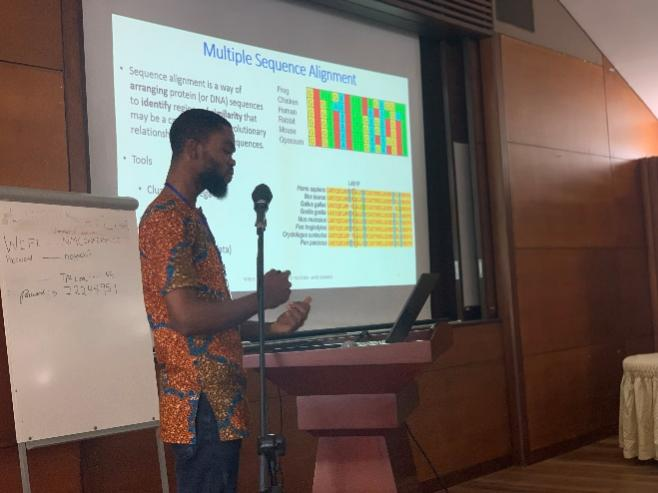
Multi-locus sequence typing (MLST) techniques were discussed by Mr. Bright Agbodzi, who also highlighted the benefits of WGS for MLST. He gave an example of how to create phylogenetic trees with web resources such as CSIphylogeny and RealPhy. Together with Mr. Amuasi and Mr. Owusu-Nyantakyi, they trained participants to align FASTA files and construct phylogenetic trees by installing MegaX software on their PCs. The usage of iToL for sketching trees was also shown by Mr. Owusu-Nyantakyi.
The CGE, CARD, PATRIC, and Pathogen Watch platforms were used to help participants identify sequence types, resistance patterns, and virulence variables. Dr. Beverly Egyir stressed the importance of these instruments in situations where the organism is known.

 DAY 5 (10TH JUNE, 2022)
Theory Session
DAY 5 (10TH JUNE, 2022)
Theory Session
Using what they learned in the workshop, participants collaborated in groups to complete the presentation slides. The purpose of the tasks and presentations was to evaluate their comprehension and application of the principles they had learnt.
Noting their development and conversations even during breaks, Dr. Beverly Egyir praised the participants for their active participation. She urged them to use and expand on the skills they had learned, pushing them to go beyond monitoring to creating diagnostic tools.
She restated the training's goals while highlighting their usefulness. Guidance in submitting sequences to NCBI for accession numbers was given by Mr. Christian Owusu-Nyantakyi.

 Presentation Session
Presentation Session
Following their 20-minute presentations, each team participated in a Q&A session. Attendee Dr. Appiah-Korang Labi, WHO technical representative, encouraged the participants to take advantage of learning opportunities and commended them for their increased professionalism and usage of terminology.
The WHO, Dr. Labi, Dr. Hedidoh, Dr. Adu, her staff, and the attendees were all praised by Dr. Beverly Egyir for their efforts. Throughout the course, she stressed the participants' dedication, growth, and collaboration. She also underlined their responsibility in using the acquired abilities to advance society.
After a group selfie and small talk, the session came to an end with attendees filling out an online survey to offer suggestions for future events.

 FACILITATORS
FACILITATORS
The training was facilitated by Dr. Beverly Egyir, (Senior Research Fellow, Department of Bacteriology, NMIMR, UG) and Dr. Bright Adu (Senior Research Fellow, Department of Immunology, NMIMR, UG).
The facilitators were supported by Ms. Felicia Owusu-Nyantakyi, Ms. Quaneeta Mohktar, Mr. Christain Owusu-Nyantakyi, Mr. Grebstad Rabbi Amuasi, and Mr. Bright Agbodzi.
























